The NINDS Intramural Program encourages scientific interactions and collaborative research projects. The NINDS supports a number of Core Facilities that provide state-of-the-art equipment and expertise to support intramural NINDS investigators. Additional NINDS-partner and NIH-wide facilities are also available to researchers. These NINDS shared core services are available to NIH employees only.
Animal Health and Care Section (AHCS) / Animal Care and Use Committee (ACUC)
The Animal Health and Care Section (AHCS) provides a comprehensive program of animal care and use for intramural investigators. The AHCS, in conjunction with the Animal Care and User Committee (ACUC), participates in assuring that the Institutes' Animal Care Program is in compliance with all applicable regulations, guidelines, and policies.
Core Facilities
Core Facilities provide shared scientific resources, cutting-edge technologies and novel approaches to support crucial research activities and accelerate scientific discovery.
|
The Bioinformatics Core Facility consists of both Bioinformatics Scientist-staff and Bioinformatics Programmer-staff. Its mission is to collaboratively assist NINDS research staff with Bioinformatics-related projects as needed. |
|
The Biospecimen Core was established in the fall of 2021, to provide sample acquisition and processing, sample tracking, storage, and sample shipment support. The core will first focus on establishing standard operating procedures, and maintaining a database on clinical information and laboratory results on each of patients from whom the specimens are acquired. For more information, please contact Niru Amin, Biospecimen Director). |
|
The Electron MIcroscopy (EM) Core Facility facilitates NINDS investigators with the opportunity to use electron microscopic techniques in their research programs. The staff provide assistance in all aspects of electron microscopy including project planning, specimen preparation and training. |
|
The Flow & Imaging Cytometry Core Facility provides intellectual, technical, training and collaborative support to NINDS and other intramural. |
|
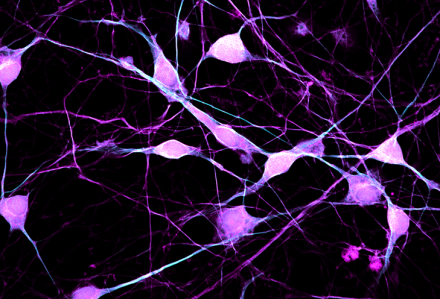
The Light Imaging Core Facility (LIF) provides intramural scientists with access to state-of-the-art light imaging equipment and expertise in light imaging techniques. |
|
The Proteomics Core Facility provides amino acid sequencing of purified proteins/peptides for NINDS investigators. The facility is also available for collaborations involving protein/peptide purification and more complicated sequencing strategies. |
|
The Clinical Proteomics Unit uses state-of-the-art mass spectroscopy based techniques to decipher the proteome in the cerebrospinal fluid and other biological materials. The search for a new Director is ongoing. |
|
The primary goal of the Quantitative MRI (qMRI) Core Facility is to provide PIs access to research MRI resources developed within and outside NINDS, and to assist in their implementation as endpoints in clinical trials of neurological diseases. |
|
The Translational Neuroscience Center (TNC) provides an infrastructure to assist researchers in pre-clinical and clinical studies to develop new diagnostics and treatments for neurological diseases. The TNC is comprised of three.
|
Clinical Trials Unit
As a part of the Office of the Clinical Director, the Clinical Trials Unit (CTU) is a group of clinical research experts who collaborate to provide NINDS intramural investigators with support in developing and executing clinical research studies, as well as regulatory guidance.
Neurology Consult Service
The NINDS intramural program provides neurological consultation services to the NIH Clinical Center with 24/7 coverage, 365 days a year, through the Neurology Consult Service. The service is staffed by neurological care providers who serve the patient care goals of the Clinical Center with diligence and attentiveness.
NINDS Partners and NIH Shared Facilities
The NIH Intramural Research Program has a long history of interactions and shared resources among investigators. Click here for more information.
Additional Resources
