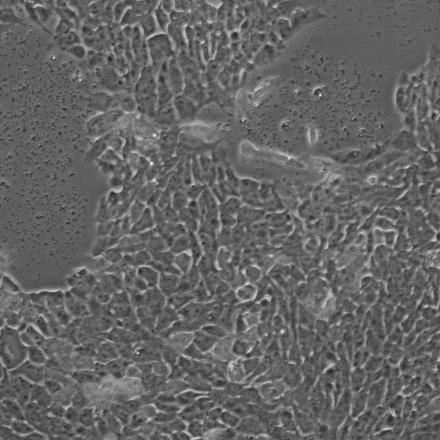
Scalable production of iPSC-derived neurons:
(3 day time lapse)
Genetic engineering of iPSCs:
New gene editing tools such as CRISPR now allow us to precisely and efficiently modify the genomes of stem cells. We use these tools to correct mutations in patient-derived iPSCs and introduce mutations into control lines, thereby creating isogenic disease model and control lines. We also use this technology to introduce transgenes into “safe harbor” loci, imparting our iPSC lines new with functionality such as doxycycline-induced differentiation, proteomic bait expression, and multi-color organelle markers for live cell imaging. Safe harbor of CRISPR machinery also allows us to use iPSC-derived neurons for genetic screens, enabling unbiased identification of neuron-specific disease-related pathways.
Proteomics:
Many of our projects leverage the power of proteomics as an unbiased platform to discover how disease-related mutations alter the biology of neurons. Because certain disease-relevant protein interactions may rely on cellular and molecular components that are found only in neurons, we use our iPSC-derived neuron system for the majority of our project. This provides an unbiased experimental platform that allows us to uncover new disease-relevant protein networks, which are then further investigated in hypothesis-driven experiments. In addition, we have modified a new enzyme-based proximity labeling proteomic pulldown technique (APEX) for use in iPSC neurons. This new approach will enables identification of transient interactions that might be missed with traditional immunoprecipitation-based proteomic experiments, and can be used to quantify the interactomes of organelles and specific bait proteins of interest.
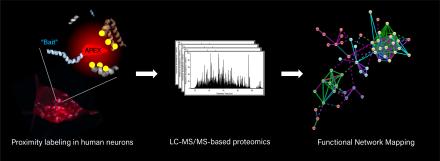
Microscopy:
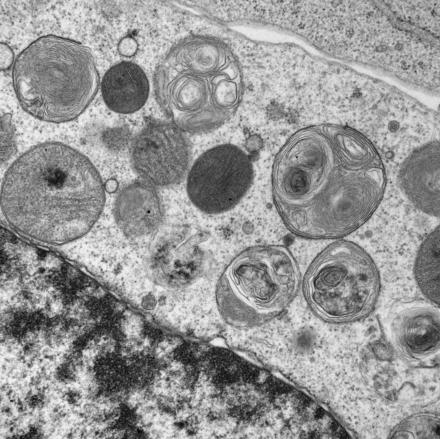
Live-cell imaging is a cornerstone of many of the hypothesis-driven experiments performed in our lab. New imaging technologies allow us to visualize protein and organelle dynamics with unprecedented spatial and temporal resolution, and new fluorescent-protein based probes enable monitoring of many cellular processes in living neurons that are relevant to neurodegeneration, such as calcium dynamics.
High Content Imaging:
In addition to high-resolution imaging, we perform high-throughput, multiwell imaging of our iPSC-derived neurons as a first step in development of large-scale phenotypic screens, which are done in collaboration with colleagues at the National Center for Advancing Translational Sciences (NCATS) and the Drug Development Unit of the Translational Neuroscience Center (TNC) at NINDS
