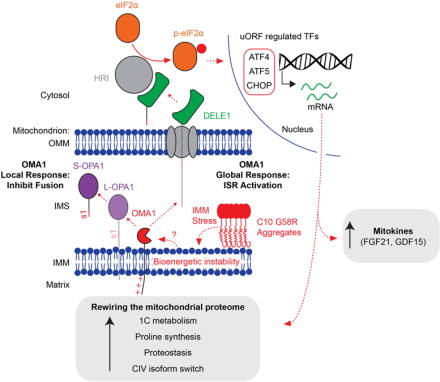
OMA1 mediates local and global stress responses against protein misfolding in CHCHD10 mitochondrial myopathy (The Journal of Clinical Investigation, 2022).
Mutations in CHCHD10 cause Isolated Mitochondrial Myopathy Dominant (IMMD), but it had been unclear which of two variants were responsible. By identifying a second family with IMMD (in collaboration with Professor Joanne Poulton (Oxford University)), we established the p.G58R mutation is the causative variant. To better understand the disease pathogenesis, we additionally generated a knock-in mouse model. In this model, the mutation caused CHCHD10 to misfold in the mitochondrial cristae of striated muscle, where it exerted a toxic effect on the inner mitochondrial membrane (IMM). We further identified that this IMM stress activates a protective mitochondrial stress response that depends on the IMM peptidase OMA1. This provided the first in vivo evidence that OMA1 mediates the mitochondrial integrated stress response, and that this response is required for survival in the setting of cristae disruption.
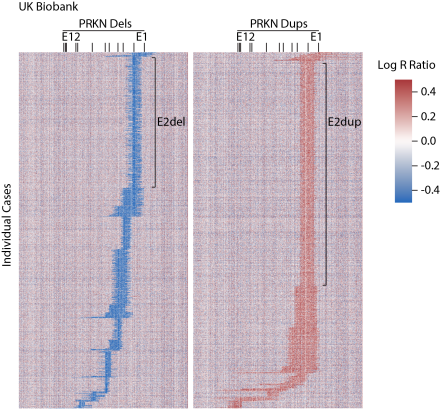
Heterozygous PRKN mutations are common but do not increase the risk of Parkinson’s disease (Brain, 2022).
Using the NIH Parkinson’s Disease cohort and large publicly available datasets (AMP-PD and the UK Biobank), we found that heterozygous PRKN mutations (coding for Parkin) are surprisingly common in the population but do not increase the risk of Parkinson’s disease. Notably, about 1% of the UK population has a rare missense variant in PRKN that is of uncertain significance, meaning that it is uncertain whether the variant is benign or pathogenic. In follow up work, we are carrying out functional studies of these and other variants in a pooled format, using fluorescent activated cell sorting together with single cell reporters of Parkin function. This work was a collaboration with the laboratory of Dr. Andrew Singleton (Laboratory of Neurogenetics, NIA).
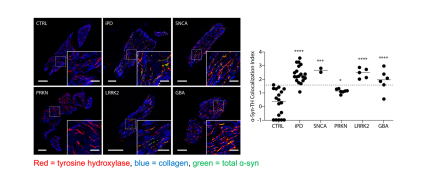
α-Synuclein Deposition in Sympathetic Nerve Fibers in Genetic Forms of Parkinson's Disease (Movement Disorders, 2021).
In collaboration with Dr. David Goldstein and Dr. Risa Isonaka (Autonomic Medicine Section, NINDS), we directly compared the extent of alpha-synuclein pathology in living patients with the most common genetic forms of Parkinson’s disease (PD). Alpha-synuclein misfolding is characteristic of idiopathic Parkinson’s disease, forming Lewy bodies in the midbrain and cortex as well as aggregates in sympathetic nerve fibers found in the skin. How alpha-synuclein deposition compares in the major genetic forms of PD, however, had been less clear. In our study, we found that patients with PRKN mutations had less alpha-synuclein deposition in biopsied skin compared to other forms of PD. This supports the view that alpha-synuclein likely does not contribute to the pathogenesis in PD caused by PRKN mutations, in contrast to other monogenic forms of PD.
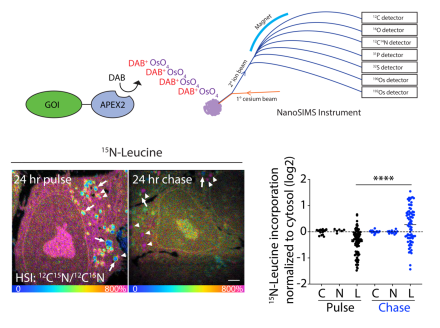
Coupling APEX labeling to imaging mass spectrometry of single organelles reveals heterogeneity in lysosomal protein turnover (Journal of Cell Biology, 2020).
Failure of local protein turnover is common in neurodegenerative disorders. In early onset Parkinson’s disease, for instance, failure of mitochondrial protein turnover by PINK1/Parkin mitophagy may lead to the degeneration of dopamine neurons. In this paper, we describe a new method that allows measurement of bulk protein turnover in single organelles by combining labelling of organelles with nanoscale metabolic imaging of protein turnover. In living cells, newly synthesized protein was metabolically labelled with stable isotope containing amino acids. Genetically encoded APEX fusion proteins were used to label the organelle of interest. The fixed cells are then analyzed using NanoSIMS, a form of secondary ion mass spectrometry, which combines high spatial resolution with accurate measurement of isotope ratios. As a proof of concept, we demonstrated for the first time that individual lysosomes in a single cell have highly heterogenous protein turnover rates. This work was a collaboration with Professor Matthew Steinhauser (University of Pittsburgh).