BG 35A RM 3D-991
35A CONVENT DR
BETHESDA MD 20892
Dr. Lucy R. Forrest received her Ph.D. in Biochemistry from the University of Oxford. Subsequent to receiving her Ph.D. degree, she carried out postdoctoral research at the Johns Hopkins University School of Medicine, at the Medical Research Council Dunn Human Nutrition Unit, Cambridge, UK and at Columbia University, New York, NY. From 2007, Dr. Forrest was Max Planck Research Group Leader at the Max Planck Institute for Biophysics in Frankfurt, Germany. In August 2013, Dr. Forrest joined NINDS, where she was granted tenure and promoted to Senior Investigator in 2017. Her laboratory at the NIH focuses on important questions relating to membrane proteins, spanning specific mechanisms of individual proteins through to general biophysical principles. Her research tools are computational and her studies are designed to inspire clear hypotheses, which are tested in close collaboration with experimental laboratories.
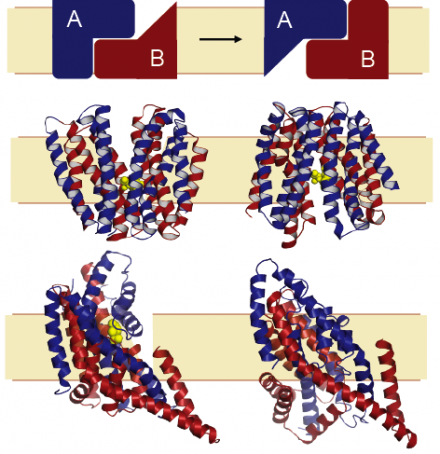
Membrane-embedded proteins are essential components of cellular organisms, allowing cells to communicate with their surroundings by providing bridges through the barrier that the lipid membrane forms. Our group is interested in understanding the mechanisms of membrane proteins using computational and theoretical approaches. Of particular interest are transporter proteins, which capture the chemical potential energy of ionic gradients (across the membrane) to facilitate movement of essential chemicals, or unwelcome toxic compounds, into and out of the cell. A fundamental question is how transporters achieve the required levels of specificity for a given chemical, or substrate, and how the protein-substrate interaction is coupled to transport of ions such as sodium. A further puzzle is how the transporter undergoes the requisite changes in shape to allow access of the substrate binding sites to either side of the membrane, while also preventing leakage (see Figure).
An essential characteristic of our work is that the hypotheses and interpretations we provide be connected with experimental evidence. Close and long-standing collaborators include the Rudnick group at Yale University, with whom we have studied transport of neurotransmitters such as serotonin. We also study glutamate transport in collaboration with the Kanner group, and acetylcholine transport with the Schuldiner lab, both from Hebrew University, Jerusalem. An ongoing collaboration with the Ziegler group at MPI Frankfurt/University of Regensburg combines computational, structural and biochemical studies on an osmolyte transporter. An aspect we find particularly fascinating is the role of repeated elements in the mechanisms of transport conformational change, which provide an elegant pseudo-symmetry and degeneracy to the system so that a single protein can adopt two pseudo-symmetric states (see Figure).
We apply a range of computational tools to understand these processes at atomic detail. To date, those techniques include sequence analysis and continuum electrostatics, as well as protein structure prediction and molecular dynamics simulation, tools for which we also develop ourselves.